On May 27, 2025, a collaborative outcome between the research groups of Professor Chen Peng and Professor Wang Chu from the College of Chemistry and Molecular Engineering at Peking University and the Peking University-Tsinghua University Joint Center for Life Sciences was published online in the journalCellunder the title "Machine-learning-assisted universal protein activation in living mice." Through sustained in-depth collaboration, the two research groups developed a protein activation technology in living organisms that integrates "machine learning and bioorthogonal cleavage reactions," achieving a breakthrough in the "proximity uncaging" strategy from living cells to living animals.
Driven by the fusion of machine learning and bioorthogonal cleavage reactions, this work yielded an innovative platform technology for in situ protein activation called CAGE-Proxvivo. This technology can transiently activate the function of target proteins in living animals and precisely regulate protein-protein interactions. Additionally, CAGE-Proxvivo can modulate cell phenotypes, induce pyroptosis in tumor cells, and enhance anti-tumor immune responses. The CAGE-Proxvivo technology has ushered bioorthogonal cleavage reactions into a new era of "arbitrary proteins + in vivo applications," marking a significant breakthrough in the development of chemical reactions in living organisms. It not only provides a powerful tool for in situ research on dynamic biological processes but also opens up new avenues for on-demand precision therapy.

Screenshot of the Paper
Bioorthogonal reactions refer to chemical reactions that can occur in biological systems without interfering with endogenous life processes. The development of such reactions has not only expanded the boundaries of biological applications for traditional organic chemical reactions but also introduced the logic of synthetic chemistry into the study of living systems, revolutionizing the molecular analysis and precise regulation of life processes and providing a new perspective for revealing complex physiological and pathological mechanisms. Bioorthogonal reactions represented by "click chemistry" solved key scientific problems such as in situ labeling and tracing of specific biomolecules in complex living systems, earning the 2022 Nobel Prize in Chemistry. However, this reaction model centered on "coupling" still has limitations and struggles to achieve the regulation and analysis of biomacromolecule functions.
The Chen Peng research group has long been committed to the development and application of "chemical reactions in living cells," pioneering the proposal and development of bioorthogonal cleavage reactions based on "bond-breaking" chemistry in the international community (Bioorthogonal Cleavage Reaction,Nat. Chem.2014, 6, 352.). Unlike traditional "linking" reactions, this cleavage reaction achieves the spatiotemporally controlled release of masking groups in target molecules through chemical bond cleavage. This strategy breaks through the technical bottleneck of using chemical reactions to regulate the functions of biomacromolecules, enabling precise "uncaging" and activation of proteins in environments such as living cells. It allows life processes and molecular interactions to be precisely initiated at specific times and spaces, opening up new avenues for functional research in living cells.
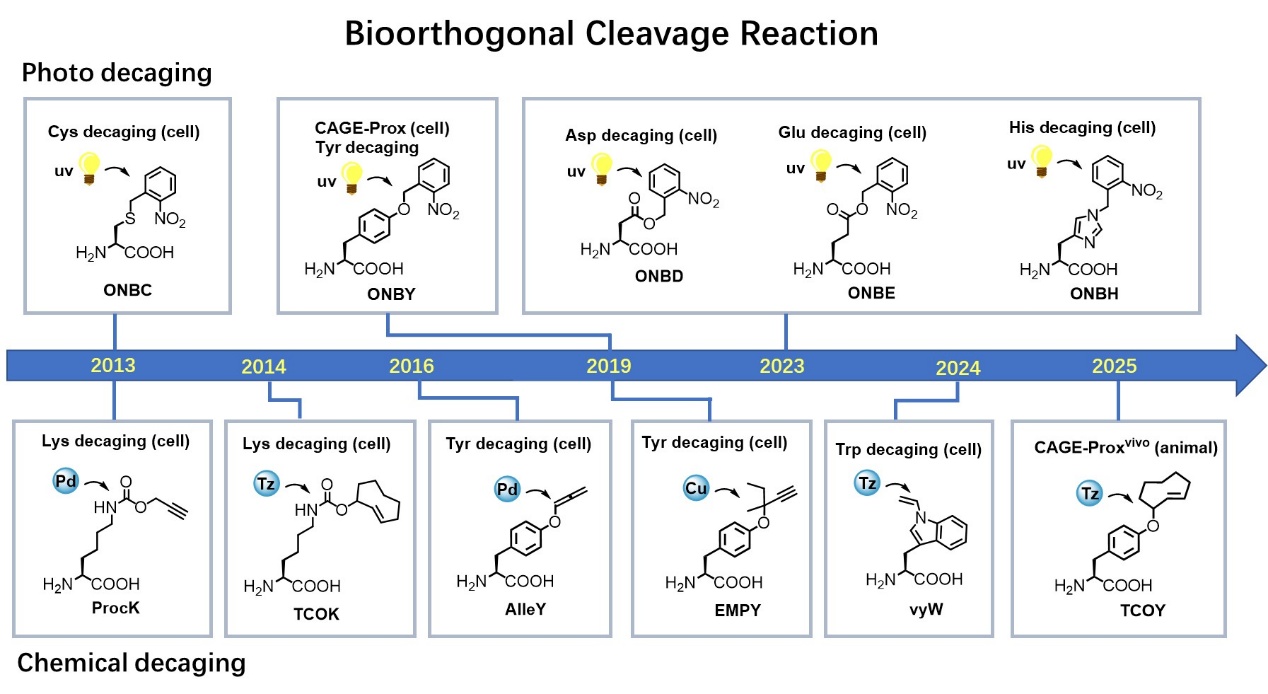
Development History of Chemical (Photo) Uncaging Bioorthogonal Cleavage Reactions
In 2019, the Chen Peng research group collaborated with the Wang Chu research group to install functional switches for proteins in living cells using UV-light-controlled bioorthogonal cleavage reactions. This strategy, known as protein "proximity uncaging" (CAGE-Prox), demonstrated strong universality and achieved transient activation of various enzyme family proteins in living cells (Nature2019, 569, 509–515).
In the work published inCell, the authors combined the inverse electron demand Diels-Alder reaction (IEDDA) and applied it to the protection and deprotection of trans-cyclooctene-tyrosine (TCOY). They trained machine learning algorithms using protein language models to capture correlations between sequence features and protein functions, supplemented by Rosetta software to simulate enzyme-substrate interactions. This improved the success rate of selecting tRNA synthetase (PylRS) variants, ultimately achieving the recognition and encoded expression of TCOY. The introduction of this "chemical switch" can temporarily shut down the function of target proteins, which can then be restored in situ in vivo through small molecule-induced bioorthogonal cleavage reactions, enabling controlled activation and obtaining a universal protein activation strategy applicable to living animals (CAGE-Proxvivo).

Machine Learning-Driven Protein Activation Strategy in Living Organisms: Training machine learning algorithms through protein language models to design and screen tRNA synthetase (PylRS) variants, achieving the recognition and encoded expression of chemically uncageable trans-cyclooctene-tyrosine (TCOY) for subsequent in vivo protein function activation and interaction regulation.
After establishing the standard CAGE-Proxvivo workflow, the authors applied it to targeted protein delivery and activation therapy. First, the authors ingeniously designed dual control of targeted delivery and chemical activation, constructing a "protein prodrug" system. Using targeted delivery, they achieved controlled, in situ activation of anthrax lethal factor, effectively inhibiting the growth of solid tumors. Building on this, the authors successfully achieved controlled-specific induction of pyroptosis within tumors using the CAGE-Proxvivo strategy. Pyroptosis, an inflammatory form of cell death mediated by Caspase, has attracted significant attention due to its potential to activate immune responses. Although traditional chemotherapeutic drugs can induce pyroptosis, tumor-specific induction of pyroptosis faces significant challenges due to low GSDME expression in many tumor cells. The CAGE-Proxvivo technology triggered Caspase-3-dependent GSDME cleavage in mouse lung cancer cells, inducing pyroptosis and triggering immune killing, effectively inhibiting the growth of secondary tumors. The authors further expanded the application range of this strategy, demonstrating the potential of pyroptosis-mediated immunotherapy in various tumor cells.
In addition, by improving the computational modeling process, the authors extended the "chemical switch" from enzyme active pockets to protein interaction interfaces, achieving precise regulation of protein-protein interactions, and developed bioorthogonal "gated" bispecific antibodies for tumor immunotherapy. In this application, the authors genetically encoded TCOY into an anti-CD3 antibody (aCD3) and combined it with a tumor-targeting module to construct a "switchable" T cell engager (TCE) with conditional activation capability. Subsequently, they restored T cell activity through in vivo bioorthogonal cleavage technology, precisely activating T cells at tumor sites, effectively avoiding systemic cytokine release syndrome (CRS) and its induced toxic reactions, and improving treatment safety. This provides a new approach to convert immunogenically "cold" tumors into highly immunogenic "hot" tumors in solid tumor immunotherapy.
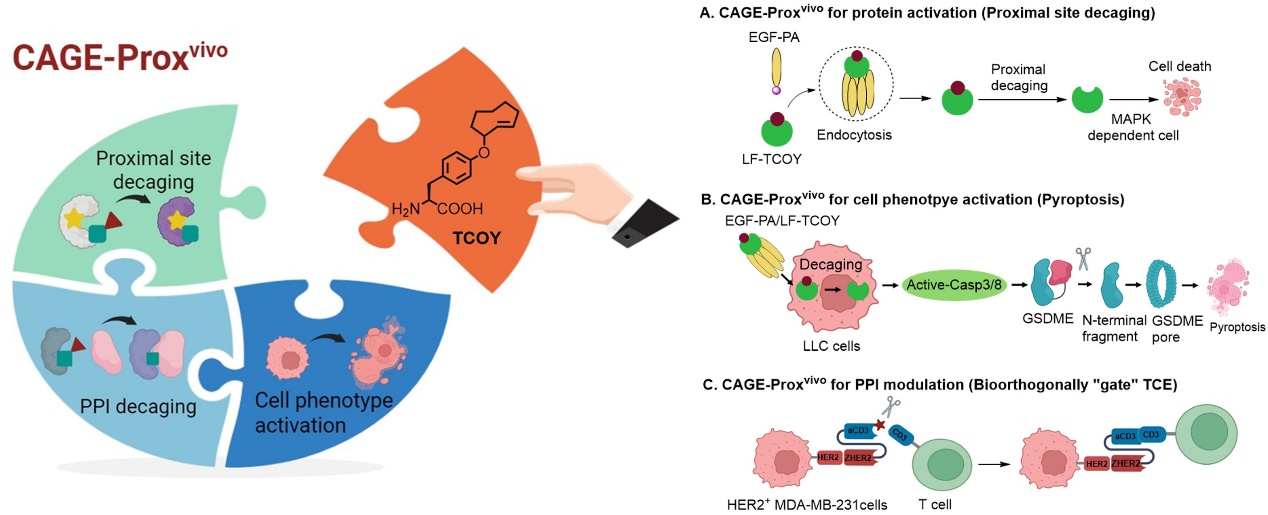
Application of the CAGE-Proxvivo Universal Platform for In Situ Protein Activation in Living Organisms: Activating protein functions on demand in living animals; regulating cell phenotypes, inducing pyroptosis in tumor cells, and enhancing anti-tumor immune responses; precisely regulating protein-protein interactions to achieve bioorthogonal "gated" bispecific antibodies for on-demand recruitment and activation of T cells.
Beyond TCOY, the authors successfully evolved PylRS variants capable of recognizing trans-cyclooctene-cysteine (TCOC) based on a unified machine learning model. Using the CAGE-Proxvivo strategy, they achieved controlled chemical masking of cysteine and precise regulation of protein functions in vivo. This research further demonstrates the effectiveness and universality of the hybrid strategy combining protein language models and structural modeling features in expanding substrate spectra, holding promise for achieving masking-uncaging of more natural amino acids and establishing a universal platform for precise manipulation of biomacromolecule functions and life processes in vivo.
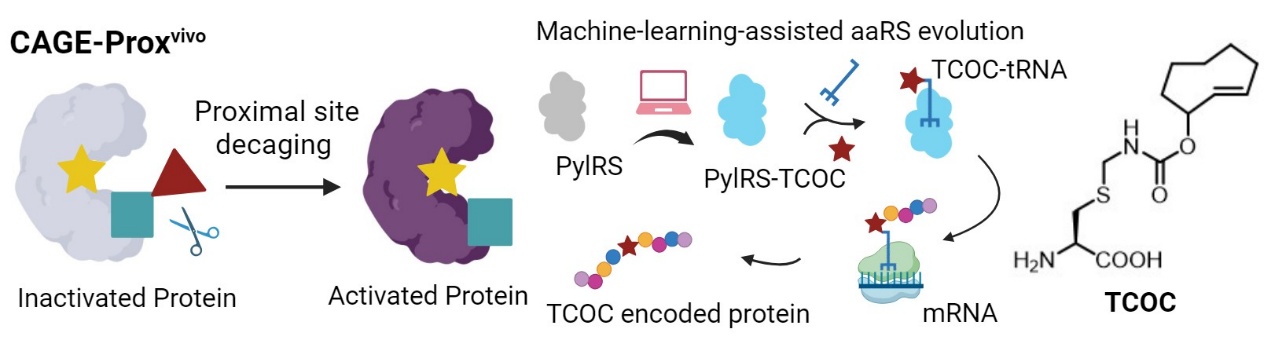
Machine learning models successfully evolved PylRS variants capable of recognizing trans-cyclooctene-cysteine (TCOC), and through the CAGE-Proxvivo strategy, achieved controlled chemical masking of cysteine and precise regulation of protein functions in vivo.
Chen Peng and Wang Chu are the co-corresponding authors of this paper. Wang Xin, a graduated doctoral student from the Academy for Advanced Interdisciplinary Studies at Peking University (currently a postdoctoral fellow at Shenzhen Bay Laboratory), and Liu Yuan, an associate researcher at the College of Chemistry and Molecular Engineering at Peking University, are the co-first authors. This work was supported by projects from the Ministry of Science and Technology, the National Natural Science Foundation of China, the Beijing National Laboratory for Molecular Sciences, the New Cornerstone Foundation, and Peking University AI4S.
